
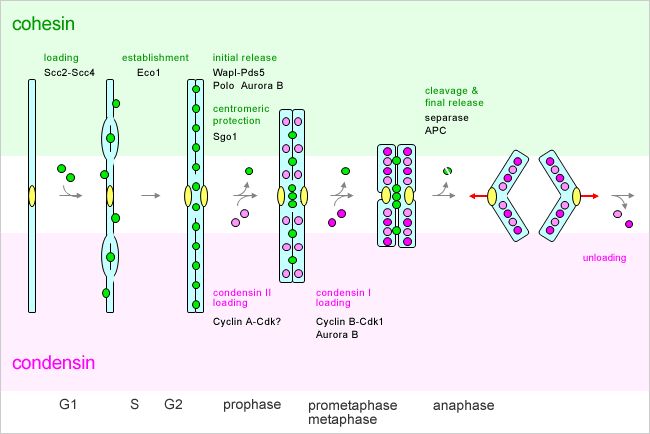
Several models of this folding have been suggested ( 3), including the ordered helical solenoid ( 4), twisted helices, and zigzag ribbon ( 5, 6). The nucleosome appears to be folded into 30 nm chromatin fibres. The structure of a nucleosome core particle has been resolved at the atomic level ( 2). 1A, a long DNA molecule with a diameter of 2 nm is wrapped around the core histone octamer, which consists of the histone H2A, H2B, H3 and H4 proteins, and forms a ‘nucleosome’, which is the most basic package of DNA ( 1). How is the 2 m of genomic DNA that is present in each human cell packaged into compact mitotic chromosomes that are 10,000 times shorter? As shown in Fig. This review article focuses on the structure of mitotic chromosomes. Chromosomes have fascinated biologists for over 100 years, since long before DNA was known to carry genetic information. The term ‘chromosomes’ was derived from the Greek for ‘coloured body’, reflecting the fact that condensed chromosomes were clearly visible with dyes under primitive light microscopes. During mitosis, chromosomes, bundles of DNA, form condensed structures to ensure the faithful transmission of the duplicated genomic DNA. This suggests that the cells in the body go through roughly 46 rounds of cell division or mitosis. The human body is made up of 60 trillion cells that originate from a single fertilized egg. Genome DNA, chromosome, condensins, scaffold INTRODUCTION Consequently, compact native chromosomes are made up primarily of irregular chromatin networks cross-linked by self-assembled condensins forming the chromosome scaffold. As a compromise to fill this huge gap, we propose a model in which the radial chromatin loop structures in the classic view are folded irregularly toward the chromosome centre with the increase in intracellular cations during mitosis. Although the classical view is that human chromosomes consist of radial 30 nm chromatin loops that are somehow tethered centrally by scaffold proteins, called condensins, cryo-electron microscopy observation of frozen hydrated native chromosomes reveals a homogeneous, grainy texture and neither higher-order nor periodic structures including 30 nm chromatin fibres were observed. Although more than 100 years have passed since chromosomes were first observed, it remains unclear how a long string of genomic DNA is packaged into compact mitotic chromosomes. ScII might thus play a role in mitotic processes such as chromosome condensation or sister chromatid disjunction, both of which have been previously shown to involve topoisomerase II.Mitotic chromosomes are essential structures for the faithful transmission of duplicated genomic DNA into two daughter cells during cell division. However, ScII appears not to be associated with the interphase nuclear matrix. ScII is a mitosis-specific scaffold protein that colocalizes with topoisomerase II in mitotic chromosomes. Analysis of the ScII B site predicted that ScII might use ATP by a mechanism similar to the bacterial recN DNA repair and recombination enzyme. ScII and the other members of the emerging family of SMC1-like proteins are likely to be novel ATPases, with NTP-binding A and B sites separated by two lengthy regions predicted to form an alpha-helical coiled-coil. ScII is structurally related to a protein, Smc1p, previously found to be required for accurate chromosome segregation in Saccharomyces cerevisiae. Here, we describe the cloning and characterization of ScII, the second most abundant protein after topoisomerase II, of the chromosome scaffold fraction to be identified.


 0 kommentar(er)
0 kommentar(er)
